
A process for human/chimpanzee divergence
Alec MacAndrew
Introduction
Can we understand what triggered the separation of the two ancestral lineages that led to human and chimpanzees today? Although it happened so long ago, scientists are homing in on the process – one which contains powerful explanatory evidence that brings a further level of detailed understanding to Human Evolution.
The most famous speciation of all
Around six million years ago, the most famous speciation of all occurred – the divergence of the lineage that eventually led to modern humans from the lineage that led to chimpanzees (1) (2) (3). That speciation initiated the evolutionary process that led to the divergence of mankind from our nearest living cousin, the chimpanzee.
There is a long-standing controversy amongst palaeontologists who debate whether modern humans evolved in Africa (the Out of Africa hypothesis), or arose simultaneously in many different places in the world after human ancestors had migrated out of Africa (the multi-regional hypothesis) (5). Recent molecular data tend to favour the Out of Africa model (genomic diversity is greater amongst Africans than populations of other continents and ranges – depending on the specific parts of the genome considered and the actual methods employed – from marginally to greatly more diverse). Although recent data support the Out of Africa hypothesis, it is not a simple, straightforward picture and there seems to have been considerable migrations into and out of Africa and Asia of Homo erectus.
However, these controversies focus on the later stages of the evolution of Homo sapiens. There is little or no disagreement that the common ancestor of humans and chimpanzees lived in Africa and that the original divergence of the two lineages took place in that continent. So whatever the more recent human evolutionary history might be, we can ask detailed questions about what caused the divergence of the two lineages from their common ancestor.
What led to the divergence of man and chimpanzee lineages?
So, if the common ancestor of man and chimpanzee lived in Africa six million years ago, what led to the genetic isolation between parts of the population that resulted in two species developing – species that were first cousins and that were the ancestors of the two lineages that eventually evolved to form modern humans and modern chimpanzees?
One hypothesis for this mechanism is that part of the population of the common ancestor became geographically isolated from the rest of the population over a long period of time, perhaps owing to a barrier such as the Rift Valley, or mountains or rivers. Such geographic isolation would prevent the process of gene flow from the population on one side of the barrier to the population on the other side. Over time, different mutations would accumulate in the two populations, resulting in different evolutionary paths and ultimately in an inability to interbreed. Once populations no longer interbreed in the wild, there can be no gene flow between them and each will follow its own divergent evolutionary path. This process is known as allopatry.
Another hypothesis is that different subgroups of the same species, for some reason such as sexual preference or specialisation to a particular narrow niche, stop interbreeding and eventually become incapable of doing so – speciation results but without geographic separation. This is known as sympatric speciation.
One type of sympatric speciation is potentially caused by major mutations that prevent successful interbreeding between parts of the population that have the mutation and parts that do not: mating between the two groups either does not result in offspring or in hybrid offspring which are themselves sterile (whilst breeding within each group – that with and that without the major mutation – is fully fertile).
It would be interesting to know which of these mechanisms led to the divergence of human and chimpanzee lineages all those millions of years ago.
Chromosomal rearrangements
So what kind of major mutation could set up a barrier to fertile mating and gene flow between subgroups within what were originally the same species? One possibility is chromosomal rearrangements. Chromosomal rearrangements occur when substantial tracts of DNA are inverted or repositioned on the chromosome. Chromosomal rearrangements that fix in the genome are relatively common. Comparison of the mouse and human genomes indicate that about 300 rearrangements have occurred since the divergence of mouse and human lineages – that is about one rearrangement every 200,000 years (5).
In the case of human and chimpanzee, the divergence occurred sufficiently long ago that there are significant numbers of rearrangements in the chromosomes but not so long that all chromosomes have rearrangements. Chromosomes 1, 4, 5, 9,12, 15, 16, 17 and 18 in humans have inversions of major tracts of code compared with homologous chromosomes in chimpanzees, and human chromosome 2 results from the end to end fusion of two acrocentric chromosomes that remain separate in all the other great apes. (6), (7), (8). Go here for a more comprehensive explanation of chromosome 2 fusion.
Could it be that some or all of these chromosomal rearrangements were responsible for triggering the speciation that separated the two lineages?
There is a simple model for the idea that chromosomal rearrangements can lead to reproductive isolation (even in geographically contiguous populations). The classical concept is that individuals that are heterozygous for the rearrangement (ie individuals who carry one copy of the chromosome that has the rearrangement mutation and one that has not) are less fertile than homozygous individuals (ie those who carry both copies of the chromosome either rearranged or in the original unmodified form). This is caused because recombination between the two copies of the chromosome in this case would result in duplication and deletion of substantial amounts of genetic code. Selection would then favour the homozygous individuals and this would lead to separation of the population into two separate homozygous subpopulations, one with and one without the chromosomal rearrangement. Those subpopulations would be, in effect, reproductively isolated, and this would eventually lead to speciation. So, as this model goes, the random mutational event of a chromosomal rearrangement can be the trigger and cause for speciation.
However, there is a major paradox with this hypothesis that calls its validity into question. If heterozygous individuals are much less fertile than normal homozygous individuals, it is hard or impossible to see how the rearrangement mutation gets established in a sub-population at all. It would simply be selected out. If, on the other hand, heterozygous individuals are as fertile or almost as fertile as homozygous individuals, the rearrangement will have no more difficulty in becoming established than any other neutral mutation. And it would present no reproductive barrier. Populations would exist that were happily polymorphic for the rearrangement (ie both rearranged and original versions of the chromosome would exist in the gene pool). In fact, populations polymorphic for chromosomal rearrangements are unknown in extant mammals, although separate populations within the same species are commonly known to carry chromosomal rearrangements.
So, we have a real problem with the simple model. On the one hand, if individuals heterozygous for rearrangements are significantly partly sterile it is impossible for the rearrangement to become established. On the other hand, if heterozygous individuals are fertile, we should expect to see substantial rearrangement polymorphism within populations, which we do not see, at least with mammals. And we are stuck without a model for human/chimp lineage speciation as the allopatric model seems inappropriate and the most likely sympatric model seems flawed.
The new model
This conundrum does have a potential solution. A more recent model of population fitness in the presence of chromosomal rearrangements points to the fact that recombination is greatly reduced in individuals who are heterozygous for rearrangements. What does this mean?
Well, recombination is a process where, within meiosis (the cell division process that creates gametes, eggs and sperm, there is random mixing of genetic material in the pairs of chromosomes, one derived from the father and one from the mother). Each individual carries two copies of each chromosome in each of its somatic cells, one from its mother and from its father. During meiosis, each pair of chromosomes exchange material so that the chromosomes in the resulting sex cells (which each have only one copy of each chromosome) contain a mixture of genetic material from the father and the mother. This process of recombination or crossing over randomises the genetic material and is a major contributor to genetic diversity.
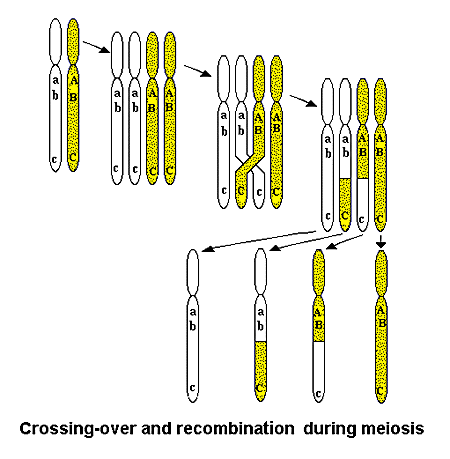
Recombination is greatly reduced in individuals that have one copy of a chromosome that is rearranged and one that is not, specifically in those chromosomes that are rearranged. Recombination of chromosomes heterozygous for a rearrangement is the main reason that heterozygous individuals were held to be partly sterile, since recombination leads to substantial deletions and duplications of genetic material resulting in unviable offspring.
But if there is little or no recombination in chromosomes heterozygous for rearrangements, then heterozygous individuals will be viable and fertile. There is no duplication or deletion of genetic material, since there is no recombination.
Since individuals heterozygous for rearrangements are fertile in the absence of recombination, rearrangements will fix with the same probability as neutral mutations (assuming the rearrangement is functionally neutral). And since there is no recombination in rearranged chromosomes, there is a barrier to gene flow in those chromosomes. If they do not recombine, then each karyotype (with and without the rearrangement) can continue to exist in the population and can continue to interbreed with viable offspring. However the mechanism for gene flow is blocked and each version of the chromosome is entirely separate and can mutate with the same degree of isolation as if the chromosome were isolated in separate species.
So we have a solution. Chromosomal rearrangements occur as a result of random mutation. Recombination is suppressed in those chromosomes allowing the mutation to fix in the population. The suppression of recombination also isolates the genetic material on that chromosome, and the chromosome evolves as though it were in a separate non-interbreeding species. Over time more chromosomes mutate and become rearranged, the lack of gene flow in the rearranged chromosomes leads to more and more divergent evolution on those chromosomes eventually resulting in sexual incompatibility between the populations that carry and do not carry the rearrangements and speciation is complete.
Predictions
If it is true that chromosomal rearrangements occurred before the speciation process was complete (or even triggered it) and that individuals with and without the rearrangement (and heterozygous hybrids) interbred fertilely for some time, what signs would that leave in the genomes of human and chimpanzee? Is there a smoking gun that we can seek as evidence for this rather neat hypothesis.
There is. Remember that during the period of interbreeding between the subpopulations, gene flow can freely occur on chromosomes that have not undergone a rearrangement (these are called co-linear chromosomes where the genes are found in the same order in both subpopulations). Gene flow, however, is restricted between the subpopulations on those chromosomes that have undergone a rearrangement (since there is no recombination between the original and the rearranged karyotype). Therefore, if there is a significant period when there is interbreeding between individuals with and without the chromosomal rearrangement (and hybrids with both the original and rearranged version of the chromosome), then one would expect to see a greater degree of divergent evolutionary mutations on those chromosomes that had undergone rearrangement compared with co-linear chromosomes. Why is this? Well, since gene flow is not restricted in co-linear chromosomes, beneficial mutations or neutral mutations linked to beneficial mutations through a selective sweep, or other mutations that fix, will tend to fix in the entire population to the same extent that they would do in any single species. On the other hand, since there is no gene flow in the rearranged chromosomes, mutations that fix in one version of the chromosome cannot spread to the other version and so every fixed mutation in either case results in a difference (divergent evolution) between original and rearranged chromosomes.
It is as though the co-linear chromosomes are in a single fully interbreeding species, and the rearranged chromosomes are already in separate non-breeding species.
Furthermore, if favourable mutations which drive the divergence of the species accumulate on those chromosomes that have rearrangements then that will lead to a signature on those chromosomes indicating the action of positive selection. This is a key prediction.
Results
So, what do we see? In a recent paper in Science (9), Navarro and Barton provide an answer. They investigated rate of protein evolution on 115 autosomal genes across the chromosomes of man and chimpanzee. The rate of protein evolution is given by a measure for each gene, Ka/Ks, where Ka is the number of non-synonymous single nucleotide substitutions between the human and chimpanzee gene and Ks is the number of synonymous substitutions. A synonymous substitution is one which codes for the same amino acid, and hence protein and which has no effect on the amino acid, protein or organism and is therefore not under selection. (Remember that there are 64 possible combinations of 3 base pair codons, coding for only 20 amino acids. There is therefore coding redundancy and each amino acid is coded for by more than one codon - so some substitution mutations are silent and have no consequence in the protein. These are synonymous mutations). A non-synonymous mutation results in a different amino acid and protein and has a potential consequence for the organism. Non-synonymous mutations, are therefore subject to selection.
First of all, the average Ks across all the genes is 1.53% and the average Ka is 0.76% (ie, in genes, only 1 in 150 nucleotides are non-synonymously different between man and chimp: or in other words only 1 in 50 amino acids differ) - this is well within the range of known coding divergence between chimpanzee and humans
Critically, the Ka/Ks ratio for rearranged chromosomes was 0.84 and for co-linear chromosomes was 0.37. This is highly significant and indicates that protein divergence is much greater in the rearranged chromosomes than in the co-linear chromosomes exactly as predicted. Furthermore if we look at the number of genes with a Ka/Ks ratio > 1, a measure that indicates positive selection at that gene, we find that genes on rearranged chromosomes have Ka/Ks > 1 in about 40% of cases whereas genes on co-linear chromosomes have Ka/Ks > 1 in only 8% of cases.
Navarro and Barton consider many potential confounding factors for these data, but find none.
They also find evidence of reduced flow of genes in rearranged chromosomes, as the hypothesis would predict. In two sets of genes selected to measure absolute K, the number of substitutions per 100 nucleotides, they find that K is greater in rearranged than in co-linear chromosomes as predicted.
In addition, a reduced rate of neutral polymorphisms in rearranged chromosomes is found. This supports the hypothesis that speciation involved the accumulation of beneficial mutations on the rearranged chromosomes, since the process of fixing those mutations would have resulted in selective sweeps that reduced the rate of neutral polymorphism on linked sites.
Conclusion
So the evidence is quite strong that the hypothesis is correct. The concept that chromosomal rearrangements initially led to chromosomal polymorphism within the breeding population. Gene flow would not occur across the original and rearranged types of the chromosome resulting in a greater evolutionary divergence on those chromosomes than co-linear ones. Indeed, those chromosomes would become a focus for beneficial mutations which were incompatible, driving the divergence of the populations and eventually resulting in total reproductive isolation and speciation. The signature of this is the greater accumulation of protein modifications on genes on rearranged chromosomes. Indeed a high (40%) percentage of genes on rearranged chromosomes are under positive selection as defined by Ka/Ks >1.
There are, of course, as Navarro and Barton point out, other possible hypotheses to explain the data including that rearrangement facilitates fixation or that rearrangements result in relaxation of purifying selection and so on. However, the data does support the hypothesis that chromosomal rearrangements fix by the period of post-rearrangement hybridisation and that rearranged chromosomes are a focus for divergent mutations which both trigger and ultimately complete the speciation.
We look forward to the publication of the draft chimpanzee genome as a great deal of light will be shed on this and many other matters concerning the evolution of man.
3rd May 2003
1. R. L. Stauffer, A. Walker, O. A. Ryder, M. Lyons-Weiler, and S. Blair Hedges, Human and Ape Molecular Clocks and Constraints on Paleontological Hypotheses, The American Genetic Association 92:469–474, 2001
2. Chen and Li, Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees, Am J Hum Genet 2001 Feb; 68(2):444-56
3. Sean Carroll, Genetics and the making of Homo sapiens, Nature 422, 849 - 857
4. On-line article by Donald Johanson, discoverer of Lucy, on the controversy between the Out of Africa and multiregional hypotheses
5. See article on chromosomal rearrangements between the mouse and human genomes here
6. Yunis, J. J., Sawyer, J.R., Dunham, K., The striking resemblance of high-resolution g-banded chromosomes of man and chimpanzee. Science, Vol. 208, 6 June 1980, pp. 1145 - 1148
7. IJdo JW, Baldini A, Ward DC, Reeders ST, Wells RA, Origin of human chromosome 2: an ancestral telomere-telomere fusion. Proc Natl Acad Sci U S A 1991 Oct 15;88(20):9051-5
8. Avarello R, Pedicini A, Caiulo A, Zuffardi O, Fraccaro M, Evidence for an ancestral alphoid domain on the long arm of human chromosome 2. Hum Genet 1992 May;89(2):247-9
9. Navarro and Barton, Chromosomal speciation and molecular divergence - accelerated evolution in rearranged chromosomes, Science 300, 321 - 324